Cornelia de Lange syndrome (CdLS) is a rare congenital disease with multisystemic involvement and developmental delay with a broad phenotypic spectrum.1–3 The classic phenotype is characterized by distinctive craniofacial features (synophrys, highly arched and/or thick eyebrows, short nasal bridge with anteverted nares and microcephaly), growth restriction with prenatal onset, intellectual disability, hypertrichosis and limb reduction defects.1,2 Atypical phenotypes are usually milder.
Sixty percent of affected individuals carry pathogenic variants of the NIPBL gene (autosomal dominant inheritance). Other involved genes are RAD21, SMC3 and BRD4 (autosomal dominant inheritance) and HDAC8 and SMC1A (X-linked inheritance). Most patients (>95%) carry de novo pathogenic variants. Haploinsufficiency is the pathophysiological mechanism.3,4
According to the guidelines of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology, variants classified as “pathogenic” and “likely pathogenic” are diagnostic of the syndrome, but “variants of uncertain significance” are not.
The involved genes are associated with the cohesin complex.1,4 This is a multifunctional complex involved in many biological pathways, most importantly in regulation of gene expression,4 so dysfunction in this complex causes global gene expression dysregulation, which explains the broad phenotypic spectrum and multisystemic involvement of the syndrome.
We present the case of a girl aged 8 years with CdLS that carried a heterozygous pathogenic variant of the NIPBL gene that had not been reported in the past.
The girl was in follow-up in the pediatric endocrinology clinic due to a history of symmetric fetal growth restriction (small for gestational age) without catch-up growth, short stature (height z score, −2.73) with delayed bone age, hypertrichosis and a peculiar phenotype (synophrys, epicanthus, anteverted nares, micrognathia and microcephaly). Pituitary and thyroid function were preserved with normal fetal development and no genital anomalies. The patient also exhibited clinodactyly of the fifth finger in both hands. She was also under multidisciplinary follow-up due to low weight, mild intellectual disability, language impairment and microcephaly.
In light of the constellation of manifestations suggestive of CdLS, with 9 points in the clinical scoring system proposed by Kline,5 in 2018 the patient was referred for testing to the department of genetics of our referral hospital. The genetic study included microarray-based comparative genomic hybridization (aCGH) and targeted exome sequencing, which yielded a negative result for CdL. Testing did also not detect any abnormalities that would explain the clinical picture.
However, given the strong suspicion of CdLS, expanded testing was requested from the reference laboratory (qGenomics) in 2023. Whole-exome sequencing identified a heterozygous splicing variant in the NIPBL gene (c.3855 + 4A > G). The interpretation was that the variant affected the exon 16 donor splice site which, despite not being the canonical splice site, could alter messenger RNA processing and give rise to an aberrant protein. The variant was not featured in commonly used variant databases and it had not been reported in any patients to date, so it was classified as “variant of uncertain significance” associated with CdLS type 1.
On the other hand, the segregation study in parental samples proved that the variant arose de novo.
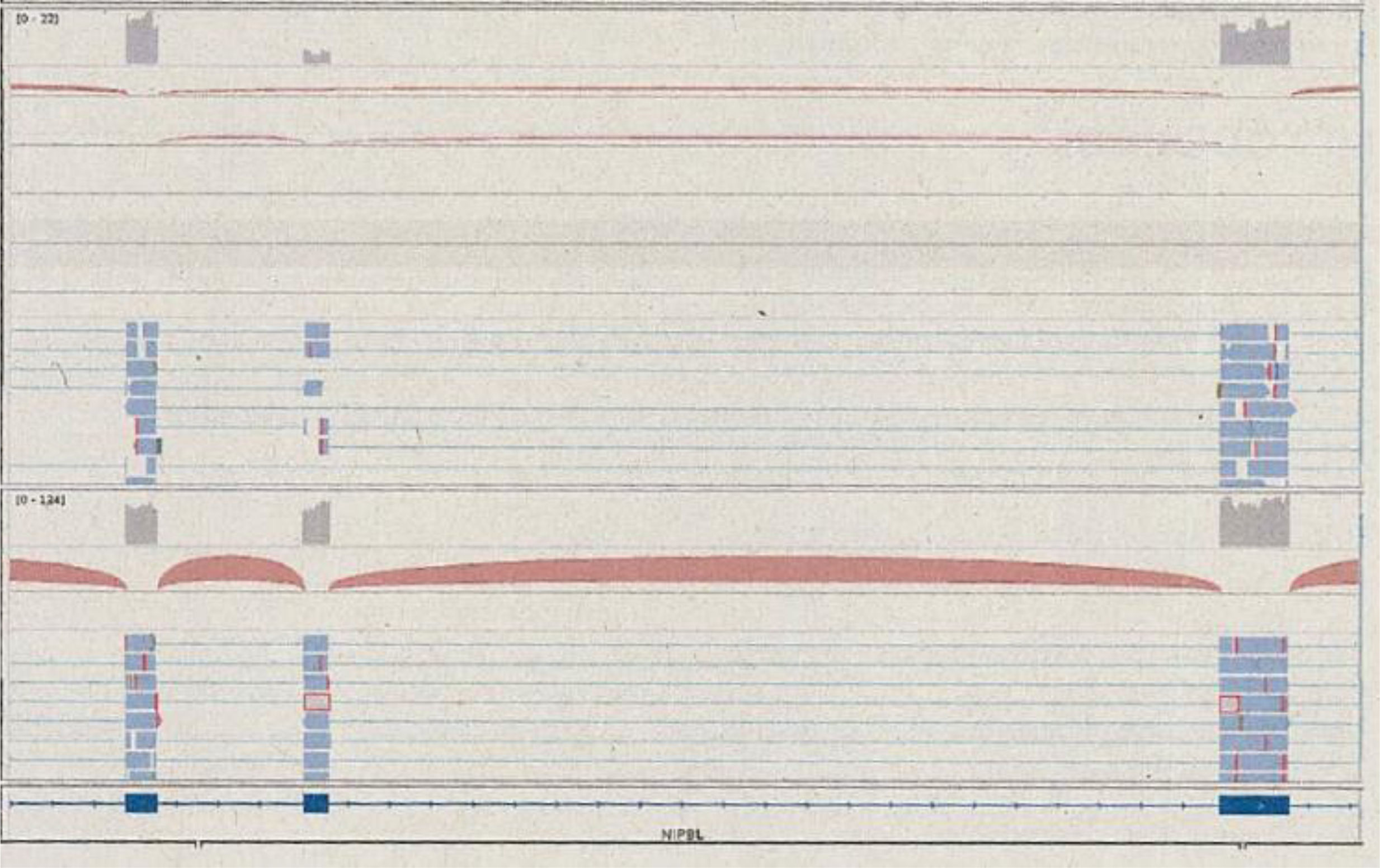
Functional assays were also conducted due to the presence of a variant of unknown significance. Transcriptome sequencing (RNA-Seq) showed that the variant did not alter the level of expression of the NIPBL gene, but affected the structure of the transcript, with skipping of exon 16 that did not disrupt the reading frame (in-frame skipping) (Fig. 1). This result supported the prediction that the variant would cause functional haploinsufficiency, compatible with the known pathophysiological mechanism of CdLS associated with NIPBL. This information allowed the reclassification of the variant as pathogenic in adherence to the guidelines of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology, and diagnosis of Cornelia de Lange syndrome 1 (CdLS-1).
Sequencing reads covering exons 15, 16 and 17. Top: cluster of reads that support the presence of the usual transcript combined with other reads that exclude exon 16 and connect exon 15 to exon 17. Bottom: results for a control, with all reads supporting the presence of the transcript of all three exons. Source: qGenomics report.
This case is relevant because the use of transcriptome sequencing (RNA-Seq) allowed the reclassification of a variant of unknown significance as a pathogenic variant and therefore diagnosis of CdLS-1 in our patient, as well as the addition of a novel pathogenic variant causing this syndrome to international genetic databases.