Natural killer (NK) cells play an important role in defense against tumor cells. The development and function of NK cells is governed by a dynamic balance between inhibition and activation of cell surface receptors, including KIR receptors.
Patients and methodA case–control study is carried out that compares a group of 46 children diagnosed with malignant diseases, the control group is made up of 82 healthy children. KIRs genes, haplotypes and ligands were determined and compared between groups.
ResultsThere are no differences in KIRs genes, KIRs haplotypes or in KIRs gene ligands between groups. However, when KIRS and ligands were jointly studied, k2DS1_C2 was significantly higher in the group of cancer children (p=0.016).
ConclusionsOur results do not provide evidence of an association between pediatric cancer disease with genotypes and groups of genes KIRs. The k2DS1_C2 genotype could predispose to susceptibility to malignant processes in children.
Las células natural killer (NK) juegan un papel importante en la defensa contra las células tumorales. El desarrollo y la función de las células NK se rige por un equilibrio dinámico entre la inhibición y la activación de los receptores de la superficie celular, incluidos los receptores KIR.
Pacientes y métodoSe realiza un estudio de casos y controles que compara a un grupo de 46 niños diagnosticados de enfermedades malignas, el grupo control está constituido por 82 niños sanos. Se determinaron y compararon entre grupos los genes, haplotipos y ligandos KIRs.
ResultadosNo existen diferencias en genes KIRs, haplotipos KIRs ni en ligandos de genes KIRs entre grupos. Sin embargo, al estudiar conjuntamente KIRs y ligandos, k2DS1_C2 fue significativamente superior en el grupo de niños oncológicos (p=0,016).
ConclusionesNuestros resultados no proporcionan evidencia de una asociación entre enfermedades oncológicas pediátricas con genotipos y grupos de genes KIRs. El genotipo k2DS1_C2 podría predisponer a la susceptibilidad a procesos malignos en la población infantil.
Advances in molecular biology have helped to provide more complete knowledge of the influence of genetic and immune factors on the pathogenesis of cancer in the paediatric age group.1 Natural killer (NK) cells, which are essential components of the innate immune system, have become one of the focuses of research.2
These NK cells act by spontaneously targeting cells considered dangerous to the host (cancer, foreign or virus-infected cells) and are therefore presumed to be key effectors in cancer immunosurveillance, transplant rejection and early viral immunity.3
The development and function of NK cells are governed by a dynamic equilibrium between inhibitory and activating cell surface receptors, including killer-cell immunoglobulin-like receptors (KIRs).4,5 The KIR genes are on chromosome 19q13.4. To date, the KIR gene family comprises 16 loci, including 2 pseudogenes (KIR2DP1 and KIR3DP1) and 4 KIR framework genes (KIR2DL4, KIR3DL2, KIR3DL3 and KIR3DP1). Fourteen functional and highly homologous KIR genes encode the key receptors that trigger the activation (KIR2DS1-5 and KIR3DS1), inhibition (KIR2DL1-3, KIR2DL5 and KIR3DL1-3) or activation and inhibition (KIR2DL4) of NK cells.5 Several studies have analysed a possible association between the KIR genes and the development of certain forms of childhood cancer, mainly leukaemias.6–12
On the basis of KIR gene content, two KIR haplotype groups, A and B, have been recognised. The group A KIR haplotype is found in all populations and comprises five KIR inhibitors (KIR2DL3, KIR2DL1, KIR2DL4, KIR3DL1, KIR3DL2) and the activating gene KIR2DS4. The group B haplotype consists of diverse genetic content including several genes (KIR2DL2, KIR2DL5, KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS5 and KIR3DS1) that are not found in the group A haplotype. Most of the KIR genes that make up haplotype B have an activating function. There have been reports of the incidence of specific KIR haplotypes or KIR genes with susceptibility to human diseases, such as autoimmune disorders,13,14 recurrent miscarriages,15,16 infectious diseases17 and cancers.18
The KIR genes are highly polymorphic and interact with the class I human leukocyte antigen (HLA) molecule, which is equally polymorphic. The KIR genes are expressed on the surface of NK cells and some T cells, and they regulate the development and function of these cells through interaction with their cognate HLA ligands,19 thereby producing a varied effect on the activity of NK cells. The expression of class I HLA alleles on the cell surface enables NK cells to be recognised as self and helps them to target non-self entities, such as cancer cells and some virus-infected cells.6,20–22
In an effort to address the genetic factors that could contribute to susceptibility to childhood cancer, we conducted a case–control study and assessed KIR genes/genotypes/HLA ligands with the aim of investigating the association between KIR genes/genotypes in children with cancer.
Materials and methodsWe documented 46 cases of children aged 0–14 years, diagnosed with and treated for malignant diseases between December 2010 and March 2017. All of them were recruited in the paediatric haematology-oncology unit of the Hospital Universitario Materno Infantil Virgen de las Nieves in Granada (Spain).
A total of 82 children with no history of malignancy or any other disease, aged between 0 and 14 years, served as controls.
Two blood samples were taken from each patient and aliquots of serum and whole blood were stored at −20°C and submitted to the Biobank Network, where the samples were processed. The genetic determinations were carried out at the Instituto de Investigación Biosanitaria de Granada.
1. HLA typing: in the 128 samples collected, the class I HLA-A*, B* and CW* and class II HLA-DRB1*, DQB1**, DQA1 and DP loci were analysed.
Genotyping was performed with the LABType SSO kit (One Lambda Inc., Canoga Park, CA, USA) using Luminex xMAP technology. Luminex is a reverse polymerase chain reaction (PCR) sequence-specific oligonucleotide (SSO) system involving the amplification of specific regions within major histocompatibility complex (MHC) I or II with group-specific primers, followed by denaturation and rehybridisation of the amplified DNA with Luminex beads covered with SSO probes to identify the presence or absence of specific alleles. The assignment of the HLA typing was based on the reaction pattern compared to patterns associated with published HLA gene sequences.
2. KIR typing: the LABType SSO genotyping kit was used for 14 KIR genes and 2 pseudogenes. Of these, 8 genes (KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL4, KIR2DL5, KIR3DL1, KIR3DL2 and KIR3DL3) have an inhibitory function and 6 (KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5 and KIR3DS1) are activating. For hybridisation, oligonucleotide probes and PCR amplification of exons 3, 4, 5, 7, 8 and 9 of chromosome 19 were used.
Quantitative data were obtained with the Luminex 200 system (Luminex Corporation, Austin, TX, USA) and were analysed using the Luminex 100 version 2.3 software, following the manufacturer's instructions.
For the haplotype study, patients with a group A KIR haplotype were considered to have a fixed set of nine genes (KIR3DL3, KIR2DL3, KIR2DP1, KIR2DL1, KIR3DP1, KIR2DL4, KIR3DL1, KIR2DS4, KIR2DL2), which include a single activating gene (KIR2DS4), two pseudogenes (KIR2DP1 and KIR3DP1) and six inhibitory genes, therefore constituting an inhibitory haplotype. Carriers of two copies of the group A KIR haplotype were considered AA genotypes, while those who lack any of the four variable genes (KIR2DL1, KIR2DL3, KIR3DL1 and KIR2DS4) were regarded as bearers of two copies of the group B haplotype (BB genotypes). All the remaining combinations were designated as heterozygous, bearing both haplogroups, that is, AB genotypes. The Bx haplotypes were characterised by the presence of additional activating genes and the absence of variable inhibitory KIR genes specific to group A (KIR2DL2, KIR2DL3 and KIR3DL1).
Statistical analysisWith regard to the study variables, the independent variable was the presence or absence of malignant disease and the dependent variables were the HLA genotype and the KIR type.
The data were managed and analysed using the software SPSS 19.0 for Windows (SPSS Inc., Chicago, IL, USA). The comparison between groups was performed with the chi-square test for qualitative variables and Student t test for quantitative variables.
The independent effect of each factor was determined using a logistic regression model.
A p value of less than 0.05 was considered statistically significant.
The data were managed and analysed using the software SPSS 19.0 for Windows (SPSS Inc., Chicago, IL, USA).
Ethics statementThis study was carried out in accordance with the most recent update of the Declaration of Helsinki. We obtained signed informed consent for the participation of all cases and controls. The project was approved by the Provincial Ethics Committee.
ResultsThe group of cancer patients was made up of a total of 29 boys (63%) and 17 girls (37%), aged between 0 and 14 years. As for the type of cancer, 38 had acute lymphoblastic leukaemia, 4 had lymphoblastic lymphomas and there were 4 with histiocytosis. Of the 82 children in the control groups, 48 were boys (58.54%) and 34 were girls (41.46%).
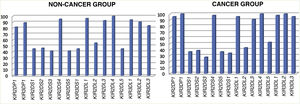
KIR genesFig. 1 shows the distribution of KIR genes in the cancer patients and the control group.
We found significant differences in the KIR pseudogenes KIR2DP1 and KIR3DP1. The P values for inhibitory KIR genes KIR3DL2 and KIR3DL3 neared statistical significance (Table 1).
Comparisons of KIR genes and HLA ligands between the group of children with cancer and the control group.
| Cancer patients (n=46) | Controls (n=82) | p | ORa (95% CI) | |
|---|---|---|---|---|
| KIR genes | ||||
| KIR2DP1 | 42 (95.5%) | 67 (81.7%) | .031 | 4.7 (1.023–21.604) |
| KIR3DP1 | 44 (100%) | 73 (89%) | .026 | |
| KIR3DL2 | 44 (100%) | 74 (90.2%) | .050 | |
| KIR3DL3 | 42 (95.5) | 69 (84.1%) | .062 | |
| HLA ligands | ||||
| C1C1 | 17 (39.5%) | 19 (23.2%) | .157 | |
| C1C2 | 19 (44.2%) | 45 (54.9%) | ||
| C2C2 | 7 (16.3%) | 18 (22.0%) | ||
| BW4 | 37 (82.2%) | 64 (78%) | .54 | |
Despite the fact that there were children in whom all the activating KIR genes were present and others with all the inhibitory KIR genes, we did not find significant differences when we compared the two groups.
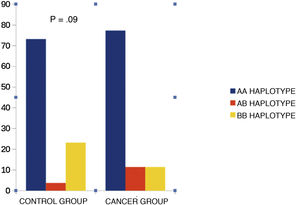
KIR haplotypesWhen we compared KIR haplotypes, we did not find differences between the groups (Fig. 2).
KIR ligandsWe then proceeded to study the KIR ligands (C1, C2, Bw4 and Bw6). The comparison between groups showed no statistical significance (Table 1).
KIR-ligand combinationsThe frequency of KIR2DS1_C2 was significantly higher in the group of cancer patients (p=.016). However, the OR of 0.489 (0.224–1.069) with 1 degree of freedom was not statistically significant (p=.07).
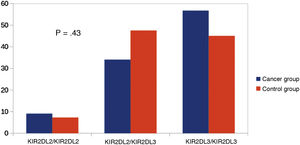
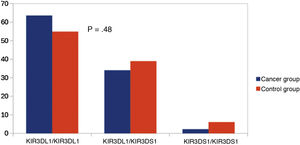
Other KIR studiesSince KIR2DL2 and KIR2DL3 are alleles of the same gene, we considered making a comparison between KIR2DL2/KIR2DL2 (homozygous for KIR2DL2), KIR2DL3/KIR2DL3 (homozygous for KIR2DL3) and KIR2DL2/KIR2DL3 (heterozygous). We did not find significant differences in this comparison (Fig. 3). Similarly, since KIR3DL1 and KIR3DS1 are alleles of the same gene, we performed an analysis comparing KIR3DL1/KIR3DL1 (homozygotic for KIR3DL1), KIR3DS1/KIR3DS1 (homozygous for KIR3DS1) and KIR3DL1/KIR3DS1 (heterozygous). There were no significant differences between the groups (Fig. 4).
DiscussionThe repertoire of KIR genes recognises specific loci of class I HLA molecules and forms a series of receptor-ligand interactions, which determines the response of NK cells.5 The interactions of NK cells depend on the combinations of KIR and HLA class I variable gene products. Activation of NK cells depends on the balance between inhibitory and activating receptors. In this study, we analysed the presence or absence of genes for 16 KIRs, including both inhibitory and activating KIRs. We also analysed KIR genes and their ligands in children with malignancies compared with a control group, as well as KIR haplotypes.
Because of the crucial role of KIRs in regulating the activity of NK cells, analyses of KIR genotypes in various childhood cancers have been performed6,7,9,10,23,24 to clarify the potential contribution of KIR variability to susceptibility to the disease and its response to treatment.
It was observed that none of the individual KIR genes, either activating or inhibitory, tended to increase or reduce the risk of developing malignant diseases in the study population. Significant differences were found only in the KIR pseudogenes KIR2DP1 and KIR3DP1. The lack of association of individual KIRs with malignancies has not been properly clarified. Previous studies have produced contradictory results in this respect: Almalte et al.25 conducted a case–control study in Canadian paediatric patients of French origin by studying the frequency of the 6 activating KIR genes and found that carrying activating KIR genes was associated with a lower risk of developing B-cell acute lymphoblastic leukaemia (B-ALL) in these children, and that the higher the number of activating KIR genes, the lower the risk of developing the disease.
Misra et al.9 support the view that the activation and inhibition of signals have cumulative effects on modulation of the effector function of NK cells in cases of childhood ALL. This study found a change towards over-activation of NK cells depending on the individual's KIR gene content, which comprised a higher number of activating genes and a lower number of inhibitory genes (KIR2DL3, KIR2DL2 and KIR2DL5) in the cases of childhood ALL compared to the controls.
In contrast, Babor et al.8 did not report any association between individual KIR genes and childhood B-ALL in children of European origin. Nor was an association found in the study by Jiang et al.6
In our study, none of the KIR haplotypes correlated with the risk of malignant disease in childhood. De Smith et al.26 found that the AA haplotype was more common in children with ALL, and this finding is also present in the study by Almalte et al.25 A study27 showed that being AA homozygous offered protection against leukaemia in adults in southern China. The NK cells of individuals homozygous for KIR A were strongly cytotoxic to leukaemia cells and the frequency of KIR activation increased in cases of childhood ALL. In the study by Misra et al.,9 persons with BB genotypes had a higher risk of childhood ALL (2.5 times) than those with AA or AB genotypes.
For combinations of specific inhibitory and activating KIR genes and their HLA ligands, our study shows an increased risk of malignant disease in the KIR2DS1-HLAC2 combination. This risk has also been highlighted previously9 and may be due to the triggering of an inappropriate localised hyperreactivity of NK cells that exacerbates the risk of growth of leukaemia cells. Although the exact significance of this finding is unknown, given that different numbers of activating KIR genes may be inherited between individuals, the frequencies of various combinations of activating KIR genes and their class I HLA ligands could modulate the risk of tumours in childhood. The contradictory reports may be influenced by the different ethnicities and different tumours analysed.
In view of these results, we recommend further investigation of the expression and function of KIR genes, HLA ligands and the expression of class I HLA molecules in tumor cells to acquire a better understanding of the mechanisms underlying the possible dysfunction of NK cells in cancer.
FundingThis study was supported by grants from the Instituto de Salud Carlos III, co-funded by the European Regional Development Fund (ERDF) (contract numbers PI12/00378, SAS-PI-0239/2012, AC-0073-2013).
Conflicts of interestThe authors have no conflicts of interest to declare.
Please cite this article as: Gómez-Luque JM, Urrutia-Maldonado E, Rueda PM, Abril-Molina A, Ocete-Hita E. Estudio de casos y controles de los receptores de tipo KIR (killer inmunoglobulin-like receptor) en oncología. Anales de Pediatrí. 2022;96:410–415.