One hundred thirty million Chlamydia trachomatis infections are reported worldwide each year. Nineteen serotypes of this pathogen can cause infection in pregnant women and neonates. The distribution of these genotypes in newborns with respiratory infections in Mexico is unknown.
Material and methodsWe tested 1062 bronchial lavage samples from neonates with respiratory distress syndrome for Chlamydia infection. The diagnosis of Chlamydia was made by plasmid detection with an in-house PCR assay, and genotypes were identified using a PCR-RFLP assay for the ompA gene.
ResultsThe genotyping of 40 strains identified 14 as I/Ia (35%), 13 as E (32.5%), 7 as D (17.5%), 5 as F (12.5%), and 1 as L2 (2.5%). The relative risk analysis showed that genotype D was associated with neonatal sepsis (RR, 5.83; 95% confidence interval [CI], 1.51–25.985; P < .02), while the I/Ia genotype was significantly associated with chorioamnionitis in the mother (2.8; 95% CI, 1.4−5.5; P < .05).
ConclusionsAlthough C. trachomatis genotypes I/Ia and E of were the strains involved most frequently in respiratory infections in Mexican neonates, 80% of patients with genotype F developed respiratory disease. In contrast, genotype D was associated with neonatal sepsis, and genotype I/Ia with chorioamnionitis.
Cada año se notifican ciento treinta millones de infecciones por Chlamydia trachomatis en todo el mundo. Diecinueve serotipos de este patógeno pueden causar infecciones en mujeres embarazadas y recién nacidos. En México se desconoce la distribución de estos genotipos en recién nacidos con infecciones respiratorias.
Material y métodosSe analizaron mil sesenta y dos muestras de lavado bronquial de neonatos con síndrome de dificultad respiratoria para detección de infección por clamidia. El diagnóstico de clamidia se realizó mediante la detección de plásmidos con un ensayo PCR interno y los genotipos se identificaron mediante un ensayo PCR-RFLP del gen ompA.
ResultadosEl genotipado de 40 cepas identificó a 14 como I/Ia (35%), 13 como E (32,5%), 7 como D (17,5%), 5 como F (12,5%) y 1 como L2 (2,5%). El análisis de riesgo relativo mostró que el genotipo D se asoció con sepsis neonatal (RR = 5,83; IC 95%: 1,51–25,985; p < 0,02), mientras que el genotipo I/Ia mostró asociación significativa con madres que desarrollaron corioamnionitis (2,8; IC 95%: 1,4–5,5; p < 0,05).
ConclusionesSi bien los genotipos I/Ia y E de C. trachomatis fueron la causa más frecuente de infección respiratoria en neonatos mexicanos, el 80% de los genotipos F produjeron este padecimiento. En cambio, el genotipo D se asoció con el desarrollo de sepsis neonatal y el genotipo I/Ia con corioamnionitis.
Chlamydia trachomatis is a gram-negative, obligate intracellular bacterium that is the most common sexually transmitted bacterium worldwide, with 131 million new cases reported in a year in adults and adolescents aged 15–49 years.1 Pregnant women can be asymptomatic for months, although they may develop symptoms at any time during pregnancy.2 During the first trimester, the presence of C. trachomatis prior to termination of pregnancy has been associated with the development of endometriosis or salpingitis in 20% of affected women.2 In the second and third trimesters, the bacterium has been associated with the development of chorioamnionitis, premature rupture of membranes (PROM), preterm birth, low birth weight and even stillbirth.2,3 After birth, newborns can develop pneumonia (5%–20%) and conjunctivitis (2%–8%).4 The genotypes that cause conjunctivitis most commonly are E and G, followed by D, F, and K.5,6 However, the evidence on which genotypes are associated with the development of pneumonia is poor and incomplete.7,8
In Mexico, the prevalence of C. trachomatis infection in pregnant women is 6.73%.9 However, the genotypes that cause this infection are unknown, although it has been proposed that the F genotype may be the most frequent based on studies in infertile women.10 When it comes to newborns, the prevalence of infection in Mexico is 11.6%, with a vertical transmission rate of 1.5% when the mother is treated with azithromycin before delivery.11 Despite the above, there are no reports on the prevalence of genotypes that cause infection in neonates, and only one study carried out by Hernández-Trejo et al.12 identified genotype D in the liver tissue of a newborn who died of unknown causes at the Instituto Nacional de Perinatología in Mexico City. The same study found that in 35.7% of neonatal death cases, C. trachomatis DNA was present in more than one organ.12
The objective of our study was to identify the C. trachomatis genotypes that cause respiratory and conjunctival infections in neonatal patients in a tertiary care facility in Mexico City.
Material and methodsBiological samplesWe conducted a cross-sectional descriptive study of 79 bronchial lavage samples from neonates managed between January 1, 2016 and December 31, 2019 in a tertiary care facility in Mexico City. The study included newborns with negative test results for respiratory infection caused by the most frequent causative pathogens of pneumonia without fever, with eosinophilia (eosinophil count > 400 cells/mm3) and with features suggestive of infection by atypical bacteria. The exclusion criteria were lack of a complete clinical history and antimicrobial treatment at the time or 72 h before sample collection. The collected samples were placed in 2-SP transport medium for C. trachomatis for preservation. Once in the laboratory, the samples were processed to extract nucleic acids.
Definitions of outcomes, signs and symptomsThe diagnosis of respiratory distress was based on the presence of nasal flaring, expiratory wheezing, sternal retraction, intercostal retraction, paradoxical respiration, polypnoea, fatigue and apnoea; monitoring evidence of hypoxia, hypercapnia, asphyxia and acidosis, a partial pressure of oxygen (PaO2) of less than 50 mmHg; central cyanosis breathing room air, or the need for supplemental oxygen to maintain a PaO2 greater than 50 mmHg; radiographic features considered typical of the clinical picture (diffuse reticulogranular pattern with air bronchogram or ground glass opacity). The differential diagnosis of respiratory distress included perinatal pneumonia, respiratory distress syndrome, transient tachypnoea of the newborn and meconium aspiration syndrome. We documented postpartum complications identified within 72 h of birth, including conjunctivitis and sepsis.
Respiratory infection was considered in newborns with a history of maternal infection, features of chorioamnionitis or PROM; who, after birth, exhibited signs of respiratory failure, rales, features suggestive of systemic inflammatory response with increases in heart and respiratory rates, fluctuations in body temperature or haemodynamic problems; with radiographic evidence of alveolar occupation with a patchy appearance and air bronchogram; features suggestive of systemic inflammatory response such as leucocytosis, leukopenia, or decreased platelets in the complete blood count; positive results in the C-reactive protein and procalcitonin tests or blood cultures, or identification of bacteria known to cause atypical pneumonia.
Approval for research involving human subjectsTesting for diagnosis of chlamydia infection in neonates was made after obtaining the signed informed consent of the parents or legal guardians. The study was approved by the Institutional Review Committee (ID 212250–3120-10607–01-14) and adhered to the ethical standards of the institution as well as the principles of the Declaration of Helsinki of 1975, as amended in 2000.
Genomic DNA extractionGenomic DNA was extracted with the phenol-chloroform method. The integrity of the genetic material was verified by 1% agarose gel electrophoresis. We measured the DNA concentration and purity (μg/mL) using a spectrophotometer (Nanodrop Biochrom™, GeneQuant™ 1300). The obtained DNA was stored at −20 °C until use.
Identification of C. trachomatisWe used a plasmid-based endpoint polymerase chain reaction (PCR) assay to identify C. trachomatis infection. To this end, a 241-bp fragment from coding region 1 (CDS1) was amplified with the primers reported by Mahony et al.,13 KL1 5′-TCCGGAGCGAGTTACGAAGA-3′ and KL2 5′-AATCAATGCCC GGGATTGGT-3′. Mahony et al. have previously described the PCR reaction, mixture and conditions.13 We used uninfected McCoy cell line DNA as a negative control and C. trachomatis DNA (ATCC: DUWC3X) as a positive control.
GenotypingWe identified C. trachomatis genotypes by restriction fragment length polymorphism analysis of PCR-amplified fragments (RFLP-PCR), as previously described.10,14 We analysed RFLP restriction profiles by means of gel electrophoresis in 12% polyacrylamide gel, and determined the genotype of the strain by comparing the results with a genetic database.
DNA sequencingWhen the RFLP assay did not identify any C. trachomatis genotype, a nested PCR was carried out. Amplification of variable segment 2 (VS-2) of the ompA gene was necessary to identify these genotypes. To do it, first, a 1000-bp fragment corresponding to the region between variable segments 1 and 2 was amplified with the primers described by Zheng et al.15 (CT1: 5′-TGA ACCAAGCCTTATGATCGACGGA-3′ and CT2: 5′-CGGAATTGTGCATTTACGTGA G-3′). Then, the amplification product served as a template for amplifying variable segment 2 using the primers CT3: 5′-ACTTTGTTTTCGACCGTGTTTTG-3′ and CT4: 5′-GATTGAGC GTATTGGAAAGAAGC -3′. These primers amplified a fragment of 416 bp. The reaction mixtures and conditions were similar in both PCR assays. The PCR reaction mix contained 1.75 mM of MgCl2, 30 pM/μL of each primer, 160 μM/μL of nucleotides, 1 U/μL of Taq polymerase (Promega), and 100 ng/μL of DNA. The PCR was processed in a PTC-100 thermocycler (MJ Research Inc) under the following conditions: denaturation at 94 °C for 5 min, followed by 30 annealing cycles at 94 °C, 1 min; 55 °C, 1 min; and 72 ° C, 1 min and a final extension step at 72 °C for 5 min. Last of all, the 416 bp product was sequenced with an ABI PRISM 310 analyser (PE Biosystems) using a BigDye DNA sequencing kit (PE Biosystems) according to the manufacturer's instructions.
Statistical analysisWe made a random selection of samples that turned positive for C. trachomatis to undergo genotype identification. The selection was obtained by generating random numbers between 1 and the total number of positive samples with a Casio FX-82MS scientific calculator (Casio Computer Co, Ltd; Tokyo, Japan).
The statistical analysis was performed with the software SPSS Statistics, version 24.0 for Windows (IBM Corp., Armonk, NY, USA), using the Fisher exact test. We have expressed the magnitude of the association between variables as relative risk (RR) with the corresponding 95% confidence intervals, applying the Student t test for normally distributed data and the Kruskal–Wallis and Mann–Whitney U tests were used for nonparametric data. We considered P values of less than .05 statistically significant.
We calculated the sample size required to detect an association between the clinical data and C. trachomatis infection in newborns using the following formula:
The number of positive samples for C. trachomatis was 363, with a margin of error of 15%, since there are plasmid-free C. trachomatis strains; the calculation showed that a selection of 39 samples would be an adequate size to assess for association.
ResultsPrevalence of C. trachomatis infection in newbornsFrom January 1, 2016, to December 31, 2019, the number of samples from neonates with respiratory distress syndrome tested for detection of C. trachomatis was 1062. The number of samples that tested positive for Chlamydia DNA was 363, which corresponded to a prevalence of 34.18%.
Of these 363 neonates, 207 were male and 156 female. The most frequent type of delivery was by caesarean section (65.3%) and 34.7% of deliveries were vaginal (P < .05). In 16.8% (61/363), there was a history of PROM and in 14.9% (54/363) of chorioamnionitis in mothers with chlamydia infection (data not shown).
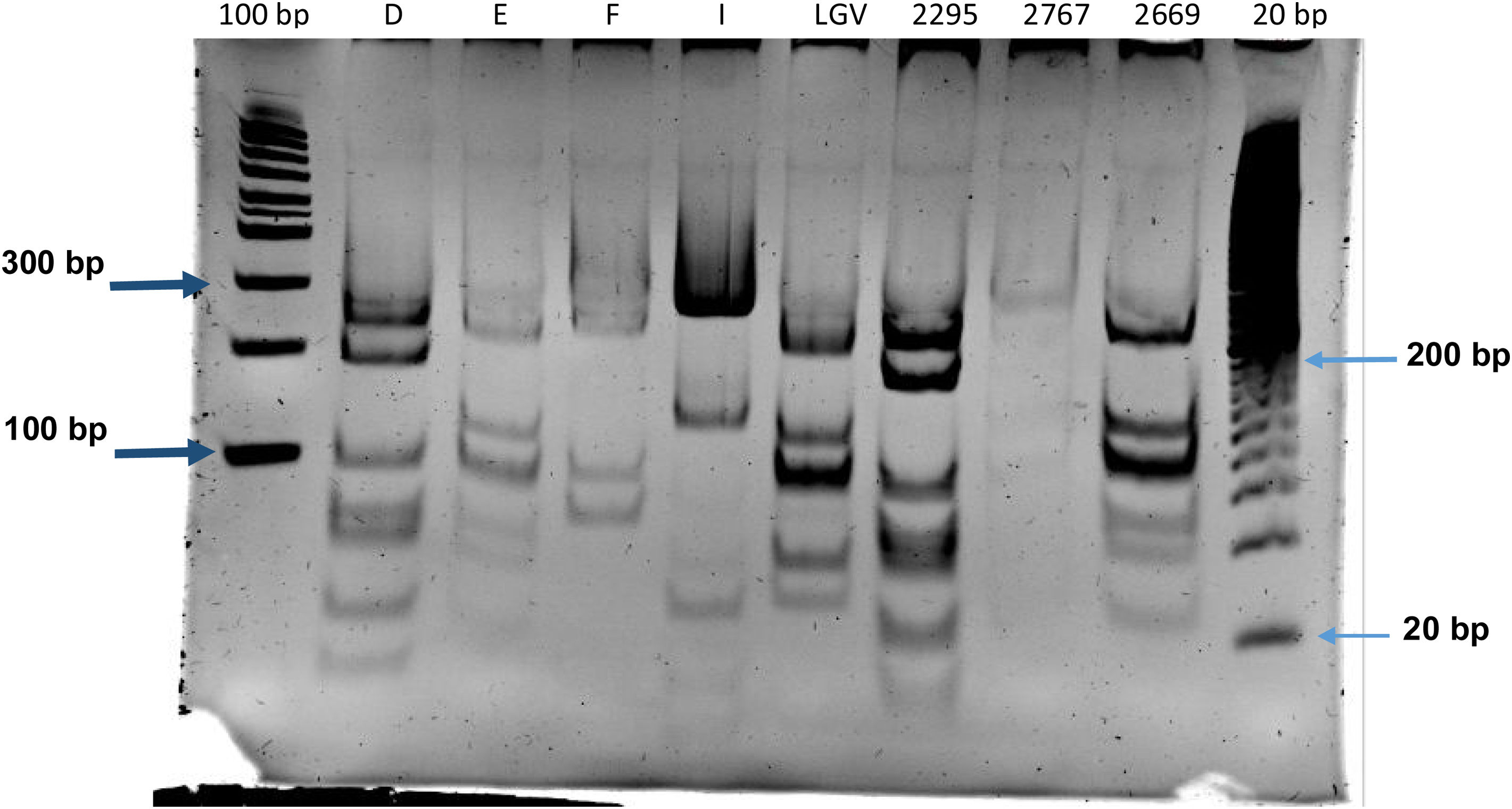
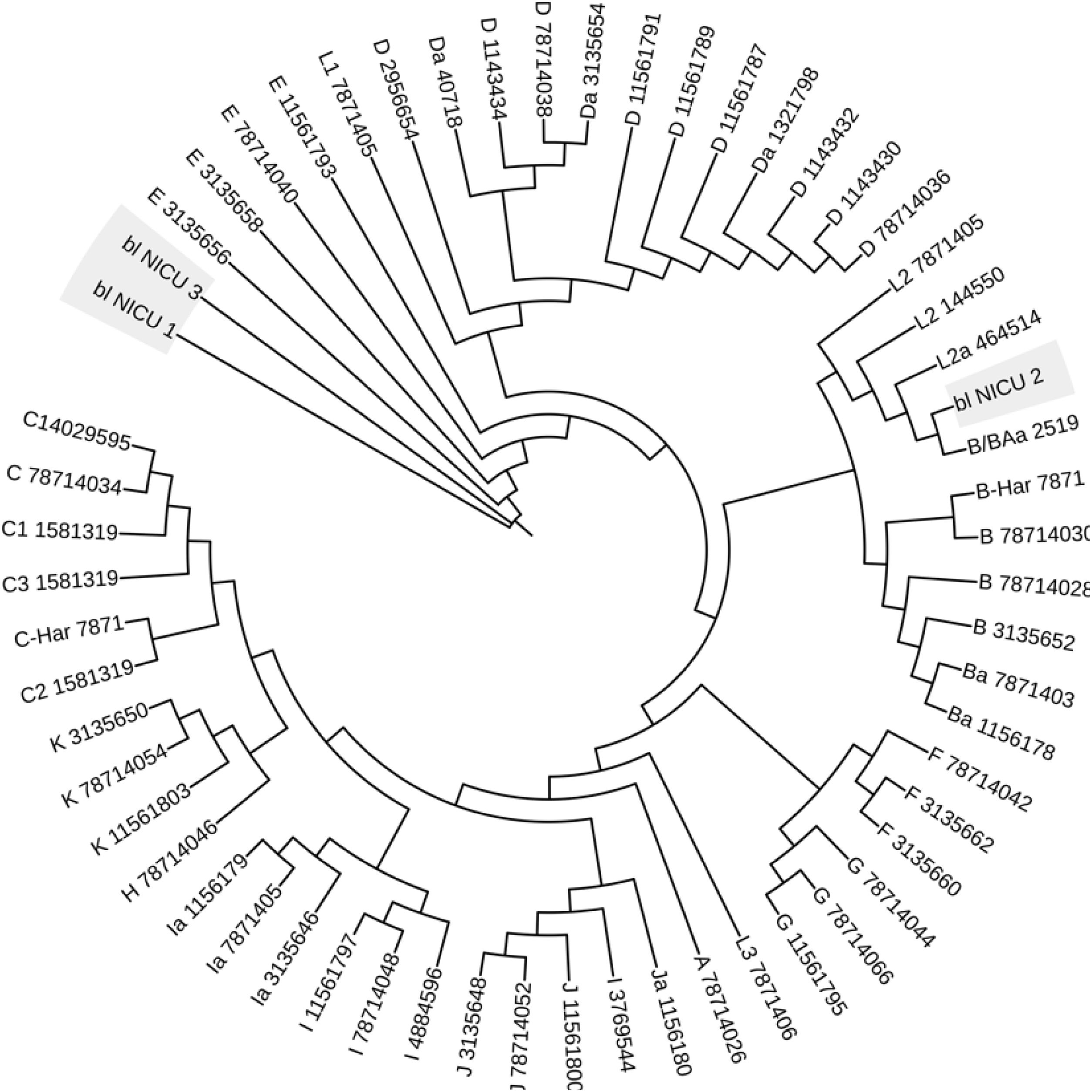
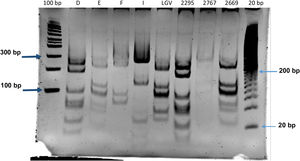
Identification of C. trachomatis genotypesOut of the total of 363 samples positive for C. trachomatis, 79 (21.8%) were randomly selected to undergo genotyping of the strain that caused the infection with the PCR-RFLP method. Fig. 1 shows the fragments obtained with the RFLP assay of Chlamydia strains and samples positive for Chlamydia. In 3 samples, the genotype could not be identified, so the nucleotide sequence of the VS-2 of the ompA gene was obtained and analysed. The resulting sequences were compared to the genotypes registered in the GenBank database of the National Center for Biotechnology Information (NCBI), and 2 of them were identified as genotype E genotype and 1 as genotype L2 (Fig. 2). The genotyping of the 79 samples only achieved identification of the genotype in 40 strains (Table 1): 14 corresponded to genotype I/Ia (35%), 13 to genotype E (32.5%), 7 to genotype D (17.5%), 5 to genotype F (12.5%), and only 1 to genotype L2 (2.5%).
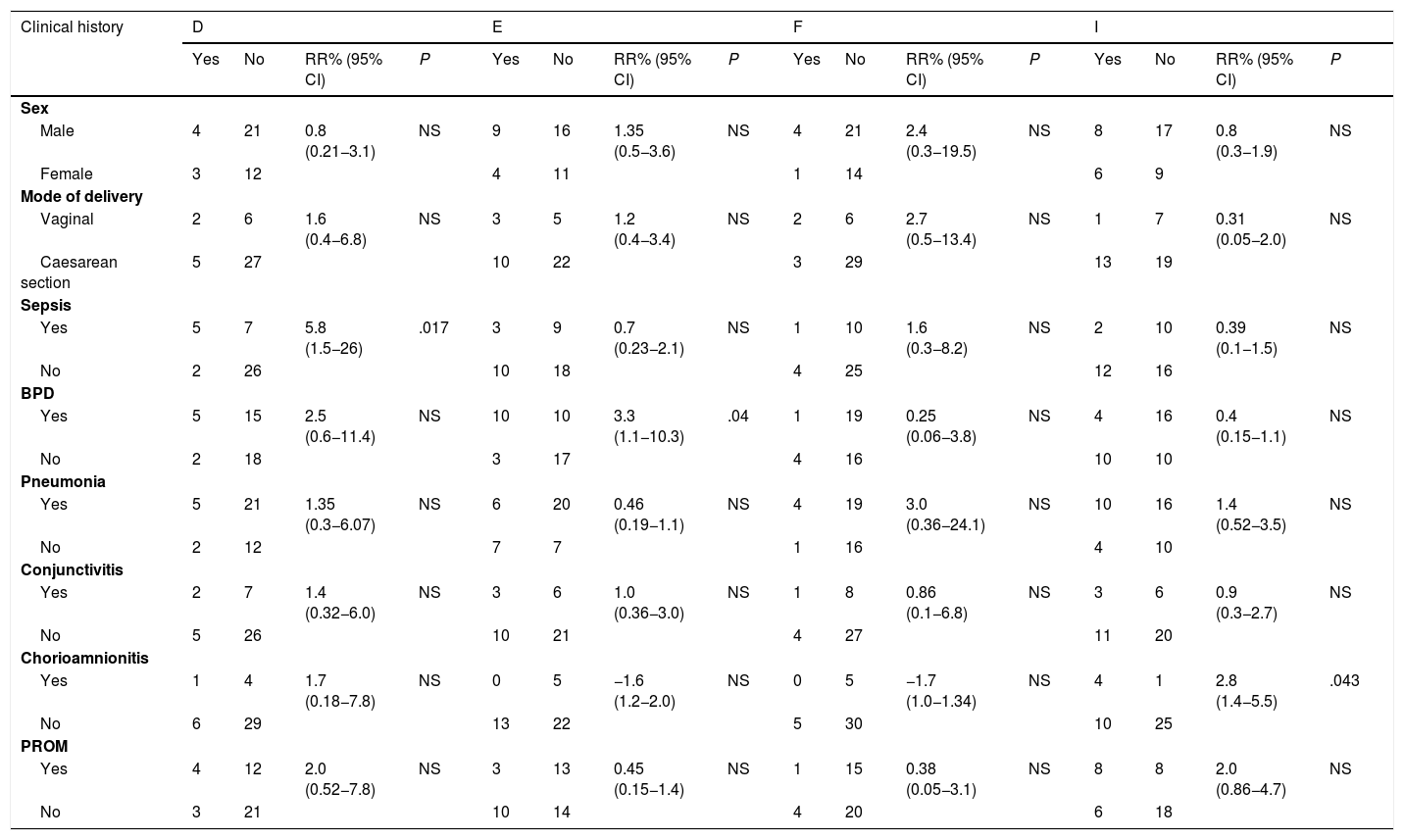
Association between Chlamydia trachomatis genotypes and clinical features of the newborn.
| Clinical history | D | E | F | I | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yes | No | RR% (95% CI) | P | Yes | No | RR% (95% CI) | P | Yes | No | RR% (95% CI) | P | Yes | No | RR% (95% CI) | P | |
| Sex | ||||||||||||||||
| Male | 4 | 21 | 0.8 (0.21−3.1) | NS | 9 | 16 | 1.35 (0.5−3.6) | NS | 4 | 21 | 2.4 (0.3−19.5) | NS | 8 | 17 | 0.8 (0.3−1.9) | NS |
| Female | 3 | 12 | 4 | 11 | 1 | 14 | 6 | 9 | ||||||||
| Mode of delivery | ||||||||||||||||
| Vaginal | 2 | 6 | 1.6 (0.4−6.8) | NS | 3 | 5 | 1.2 (0.4−3.4) | NS | 2 | 6 | 2.7 (0.5−13.4) | NS | 1 | 7 | 0.31 (0.05−2.0) | NS |
| Caesarean section | 5 | 27 | 10 | 22 | 3 | 29 | 13 | 19 | ||||||||
| Sepsis | ||||||||||||||||
| Yes | 5 | 7 | 5.8 (1.5−26) | .017 | 3 | 9 | 0.7 (0.23−2.1) | NS | 1 | 10 | 1.6 (0.3−8.2) | NS | 2 | 10 | 0.39 (0.1−1.5) | NS |
| No | 2 | 26 | 10 | 18 | 4 | 25 | 12 | 16 | ||||||||
| BPD | ||||||||||||||||
| Yes | 5 | 15 | 2.5 (0.6−11.4) | NS | 10 | 10 | 3.3 (1.1−10.3) | .04 | 1 | 19 | 0.25 (0.06−3.8) | NS | 4 | 16 | 0.4 (0.15−1.1) | NS |
| No | 2 | 18 | 3 | 17 | 4 | 16 | 10 | 10 | ||||||||
| Pneumonia | ||||||||||||||||
| Yes | 5 | 21 | 1.35 (0.3−6.07) | NS | 6 | 20 | 0.46 (0.19−1.1) | NS | 4 | 19 | 3.0 (0.36−24.1) | NS | 10 | 16 | 1.4 (0.52−3.5) | NS |
| No | 2 | 12 | 7 | 7 | 1 | 16 | 4 | 10 | ||||||||
| Conjunctivitis | ||||||||||||||||
| Yes | 2 | 7 | 1.4 (0.32−6.0) | NS | 3 | 6 | 1.0 (0.36−3.0) | NS | 1 | 8 | 0.86 (0.1−6.8) | NS | 3 | 6 | 0.9 (0.3−2.7) | NS |
| No | 5 | 26 | 10 | 21 | 4 | 27 | 11 | 20 | ||||||||
| Chorioamnionitis | ||||||||||||||||
| Yes | 1 | 4 | 1.7 (0.18−7.8) | NS | 0 | 5 | −1.6 (1.2−2.0) | NS | 0 | 5 | −1.7 (1.0−1.34) | NS | 4 | 1 | 2.8 (1.4−5.5) | .043 |
| No | 6 | 29 | 13 | 22 | 5 | 30 | 10 | 25 | ||||||||
| PROM | ||||||||||||||||
| Yes | 4 | 12 | 2.0 (0.52−7.8) | NS | 3 | 13 | 0.45 (0.15−1.4) | NS | 1 | 15 | 0.38 (0.05−3.1) | NS | 8 | 8 | 2.0 (0.86−4.7) | NS |
| No | 3 | 21 | 10 | 14 | 4 | 20 | 6 | 18 | ||||||||
BPD, bronchopulmonary dysplasia; CI, confidence interval; NS, not significant; PROM, premature rupture of membranes; RR, relative risk.
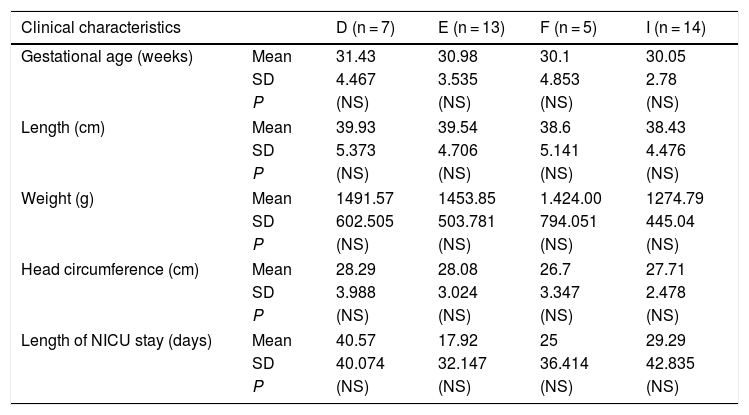
Of the 40 out of 363 newborns (11%) in whom the C. trachomatis genotype was identified, 25 were male and 15 female, and 8 had been delivered vaginally and 32 by caesarean section (Table 1). The mean gestational age at birth was 30.6 weeks (standard deviation [SD], 3.87) and the mean height, weight and head circumference were 39 cm (SD, 4.9), 1411 g (SD, 586.4) and 27.7 cm (SD, 3.21), respectively (Table 2). We did not find a significant association between any of these variables and the C. trachomatis genotype involved in the infection. Similarly, there were clinical manifestations, such as conjunctivitis or pneumonia, that were not associated with some of the identified C. trachomatis genotypes. However, we found a 4-fold increase in the relative risk of sepsis in newborns infected with the D genotype (RR, 5.83; 95% CI, 1.51–25.985; P < .02). Also, bronchopulmonary dysplasia developed more frequently in newborns infected with the E genotype (RR, 3.3; 95% CI, 1.1−10.3; P < .05), and the I/Ia genotype was significantly associated with chorioamnionitis in the mother (RR, 2.8; 95% CI, 1.4−5.5; P < .05).
Differences in somatometric measurements and clinical variables in newborns with respiratory distress and infection between different Chlamydia trachomatis genotypes.
| Clinical characteristics | D (n = 7) | E (n = 13) | F (n = 5) | I (n = 14) | |
|---|---|---|---|---|---|
| Gestational age (weeks) | Mean | 31.43 | 30.98 | 30.1 | 30.05 |
| SD | 4.467 | 3.535 | 4.853 | 2.78 | |
| P | (NS) | (NS) | (NS) | (NS) | |
| Length (cm) | Mean | 39.93 | 39.54 | 38.6 | 38.43 |
| SD | 5.373 | 4.706 | 5.141 | 4.476 | |
| P | (NS) | (NS) | (NS) | (NS) | |
| Weight (g) | Mean | 1491.57 | 1453.85 | 1.424.00 | 1274.79 |
| SD | 602.505 | 503.781 | 794.051 | 445.04 | |
| P | (NS) | (NS) | (NS) | (NS) | |
| Head circumference (cm) | Mean | 28.29 | 28.08 | 26.7 | 27.71 |
| SD | 3.988 | 3.024 | 3.347 | 2.478 | |
| P | (NS) | (NS) | (NS) | (NS) | |
| Length of NICU stay (days) | Mean | 40.57 | 17.92 | 25 | 29.29 |
| SD | 40.074 | 32.147 | 36.414 | 42.835 | |
| P | (NS) | (NS) | (NS) | (NS) | |
NICU, neonatal intensive care unit; NS, not significant; SD, standard deviation.
C. trachomatis has been associated with various conditions that affect pregnancy, foetal development, and the neonate,2,16,17 and it can contribute to preterm birth, chorioamnionitis, PROM and even neonatal death, although the latter is still under debate. In our study, the mean gestational age of the 363 neonates who tested positive for Chlamydia infection was 31.5 weeks (SD, 4.1), and there was a history of maternal chorioamnionitis in 15% and of PROMP in 17%.
Prematurity and low birth weight are commonly associated with C. trachomatis infection,3,16,17 as corroborated in our study, as most neonates were born before 32 weeks of gestation and had low birth weights. Preterm birth has been identified as the most important cause of perinatal morbidity and mortality worldwide, accounting for 27% of the nearly 4 million neonatal deaths reported annually. Chronic lung disease and infection are among the most important risk factors for perinatal mortality. The mean neonatal mortality rate reported by the Instituto Nacional de Perinatología (INPer) in this 4-year research period was of 19.9 per 1000 live births. In our study, there were 13 deaths among the 363 newborns with Chlamydia infection (3.6%) (the genotype was only identified in 1 case, and was genotype E). This suggests that C. trachomatis may be an important factor causing perinatal mortality, as other authors have proposed.18
The global prevalence of C. trachomatis infection in pregnant women ranges between 6.7 and 18%.16–21 In Mexico, it is 4.7% in pregnant adolescents and 2% in pregnant adult women.9 The reported frequency of neonatal infection ranges from 2% to 40% globally,6,22,23 and from 11.6% to 45% in Mexico.11,24 In our study, we found a prevalence of C. trachomatis infection of 35%.
It has been reported that 30%–50% of babies with chlamydia infection develop conjunctivitis and 10%–20% pneumonia.2–4 However, in our study, conjunctivitis was not frequent, developing only in 34 of the 363 infected infants (9.4%). The genotype was only identified in 9 of these cases (2.5%), and the most frequent genotypes were E (0.8%) and Ia (0.8%) (3 cases each).
The frequency of pneumonia was slightly higher, of 24.8% (90/363). The genotype Ia was identified most frequently among these cases (2.75%), followed by genotypes E (1.4%), F (1.1%) and D (1.1%). It is worth noting that 4 of the 5 patients in whom genotype F was identified developed pneumonia.
The average length of stay of these babies was 55 days, and they received a diagnosis of respiratory infection caused by atypical microorganisms. The high average length of stay could be due to complications intrinsic to prematurity and the disease itself. Only 65% of the patients had bronchopulmonary dysplasia (BPD). Carrera-Muiños et al.23 reported similar results in terms of the length of stay in neonates with Chlamydia infection.
In Mexico, there is no previous research on the genotypes of C. trachomatis that cause infection in newborns. Previous studies have found that genotype F is most frequent in infertile women and men whose partners are sterile in Mexico City.10,25 In Guadalajara, Mexico, genotypes E, F, and D are most frequent in infertile women, whereas F and E are more common in pregnant women.21 In our study, of the 40 samples in which the genotype of Chlamydia that caused infection in newborns was identified, the most frequent genotype were I/Ia (35%) and E (32.5%), followed by D (17.5%), F (12.5%), and L2 (2.5%).
Although in this study it was not possible to demonstrate that any C. trachomatis genotype was the sole cause of a condition, certain trends could be identified. Genotype D has already been reported in Mexico as the cause of neonatal sepsis. This genotype was detected in a liver sample from a deceased newborn who had features of neonatal sepsis without bacterial isolation and whose mother had had chorioamnionitis and PROM of 8 h duration.12 In this study, genotype D was 4.6 times more frequent in newborns that developed sepsis (P < .04). Sepsis by C. trachomatis is not something new; in 2016, a study conducted in Ghana found a 33.7% prevalence of sepsis and premature rupture of membranes (P < .001) in newborns of mothers infected with C. trachomatis.26 However, the authors did not identify the genotype that caused this condition.
At the international level, in Argentina, genotype D has been identified in 5.3%–7.9% of cases of neonatal conjunctivitis,5 whereas in Chile and China, it was more frequent in cases of neonatal pneumonia, with proportions of 11.8% and 12.1% of cases, respectively.7,8 In our study, these diseases were present in 22.2% and 17.4% of cases, respectively.
Genotype D is one of the most invasive genotypes because it has a cytotoxin that can lyse the cells of the genitourinary mucosa, as demonstrated in a study conducted by Stephens et al.27 This cytotoxin is encoded by genetic sequences CT165 to CT168, which produce a protein with significant homology in its amino acid sequence to that of Clostridium difficile cytotoxins.28 In addition, it causes changes in the cytoskeleton of the host cells due to its glycosyltransferase activity, which modifies the intracellular regulatory molecules, such as GTP-binding proteins of the Rho/Rac family.29–31 These cytoskeletal changes can result in loss of cell membrane integrity and thus promote the development of sepsis.
One salient finding of our study was the identification of a nucleotide sequence corresponding to a lymphogranuloma venereum (LGV) strain (C. trachomatis L2 strain) in a neonatal sample. The mother of this infant did not develop any signs or symptoms of this disease. The prevalence of L2 in newborns has not been not established; there is only one report by Beem et al.,32 who detected antibodies against serotype L1 of C. trachomatis in 18 out of 20 newborns with pneumonia. In addition, several studies have reported infection by the L2 genotype in heterosexual and homosexual men and in women who did not develop LGV,10,33 suggesting that this is an L2 variant, possibly the L2f variant. The L2 variant nucleotide sequences obtained in our study evinced the presence of deletions, substitutions and insertions in the VS1 and VS2 regions of the ompA gene.
On the other hand, genotype E is the most prevalent in neonates with pneumonia in Chile (47.1%)7 and China (42.4%)8 and in neonates with conjunctivitis in Argentina (72.4%)5 and Hungary (48%).6 In our study, genotype E was the second most frequent in neonates with pneumonia (21.7%) and identified in a single case of neonatal conjunctivitis (11.1%). However, we observed a trend in which 42.9% of infants infected with this genotype developed BPD. However, the aetiology of BPD is multifactorial, and potential contributing factors include prolonged mechanical ventilation, high concentrations of inspired oxygen, chorioamnionitis or sepsis or the degree of prematurity, among others. Further research is required to determine whether the E genotype is associated with a higher degree of pulmonary prematurity, as in this study in was not found to be associated with the development of sepsis or chorioamnionitis. In Argentina, genotype E was involved in 7.9% of cases of neonatal conjunctivitis.5 In our study, 33.3% of infants with this genotype had this condition.
As regards the F genotype, a lower frequency of infection was found in newborns compared to infertile women and men managed at the Instituto Nacional de Perinatología in Mexico City.10,25 In Chile and China, studies have reported proportions of cases of pneumonia caused by C. trachomatis corresponding to this genotype of 17.6% and 42.4%, respectively.7,8 In our study, we found a frequency of 17.4%, similar to the one in Chile. However, it should be noted that of the 5 neonates infected with the F genotype, 4 of them had pneumonia (80%), which makes this genotype particularly interesting as the possible main cause of pneumonia. In this study, we were unable to evince an association with pneumonia, possibly due to the low number of samples that tested positive for this genotype. Conjunctivitis caused by genotype F has not been described frequently in neonates.5,6 Our findings were consistent with this.
When it comes to genotype I/Ia, a prevalence of 1.3% has been reported in Argentina in neonates with conjunctivitis.5 In our study, we found genotype I/Ia in 33.3% of infants with this condition. There are no previous reports of the frequency of genotype I/Ia as the cause of pneumonia. In our study, we found this genotype in 43.5% of newborns with pneumonia.
Another interesting finding of our study was that genotype I/Ia was associated with a statistically significant 2.8-fold increase in the relative risk of chorioamnionitis; there are no previous reports on the prevalence distribution of genotypes that cause chorioamnionitis. Still, Geisler et al.34 described an association of I/Ia genotype with African American ancestry in women in the United States, with a prevalence of 16%, while a prevalence ranging from 4.2% to 6% has been reported in infertile Mexican women.10 Looking at other countries, the reported prevalence is 4.8% in Senegal, 2.1% in Costa Rica and 16% in Guadeloupe, in the Caribbean region.35–37
ConclusionAlthough genotypes I/Ia and E of C. trachomatis were most frequently involved in respiratory infections in Mexican neonates, 80% of patients with genotype F developed respiratory disease. In contrast, genotype D was associated with neonatal sepsis, and genotype I/Ia with chorioamnionitis.
Conflicts of interestThe authors have no conflicts of interest to disclose that are relevant to this article. The authors alone are responsible for the content and writing of the article.